Phylogenetic insights into Ebola dynamics in the unobserved reservoir
JT McCrone
Fred Hutch Cancer Center
May 22, 2024
Combi Seminar Series
UW - Foege Auditorium
Realtime outbreak investigation
Phylogenetics reveals underlying transmission process
Phylogenetic methods in pandemic-sized datasets
Tractable phylogenetics for the pandemic
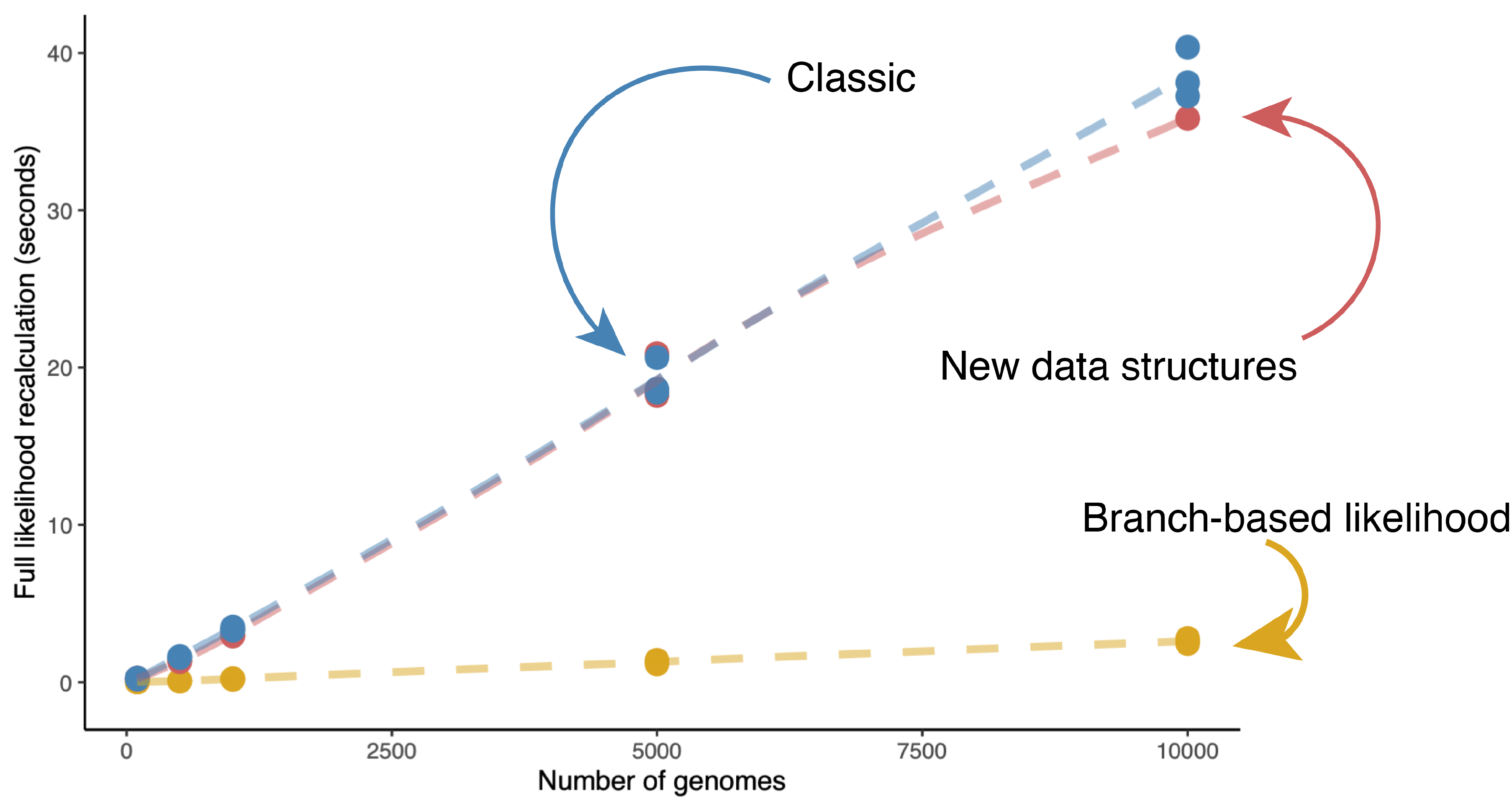
Computational time
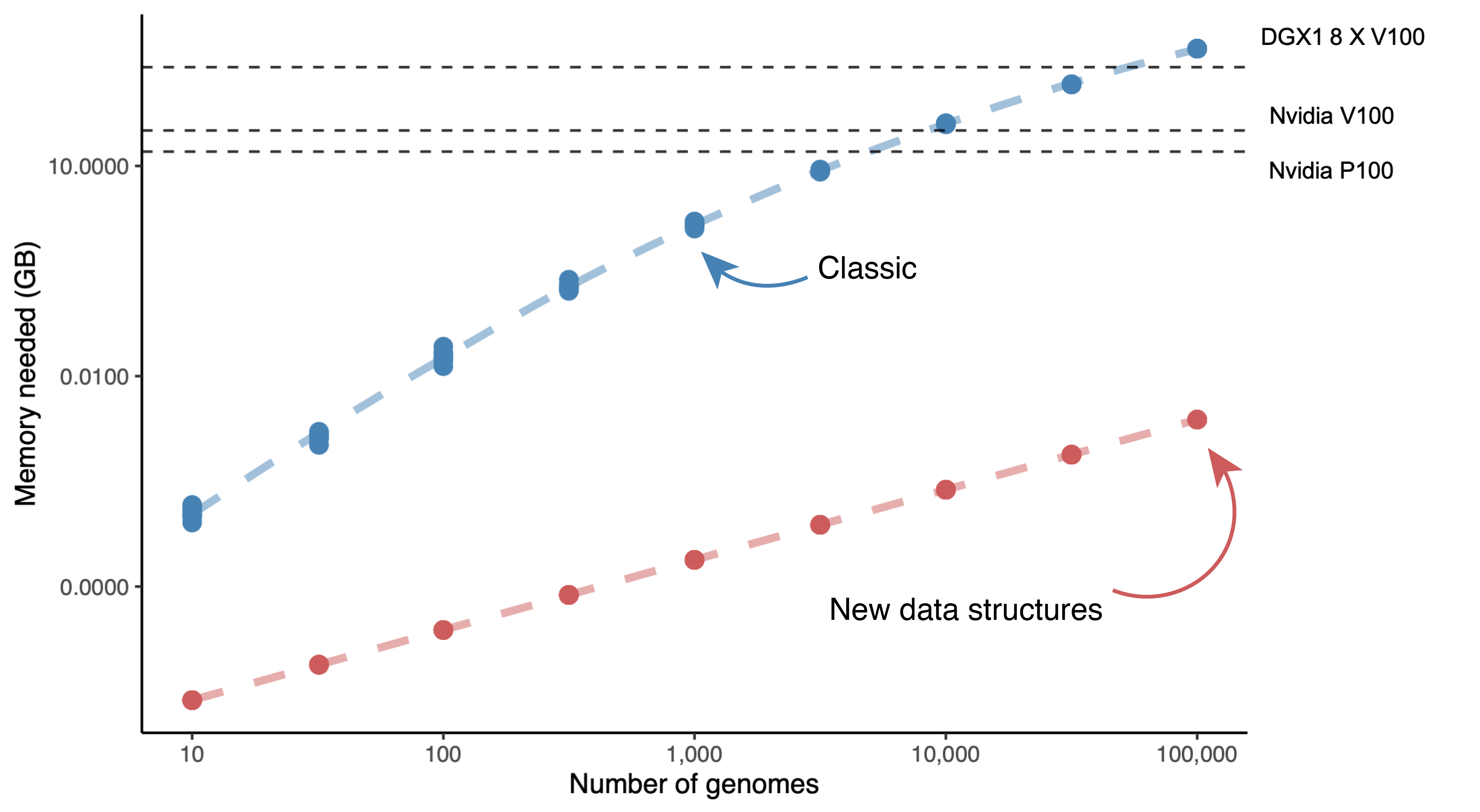
Memory usage
Within-country spread
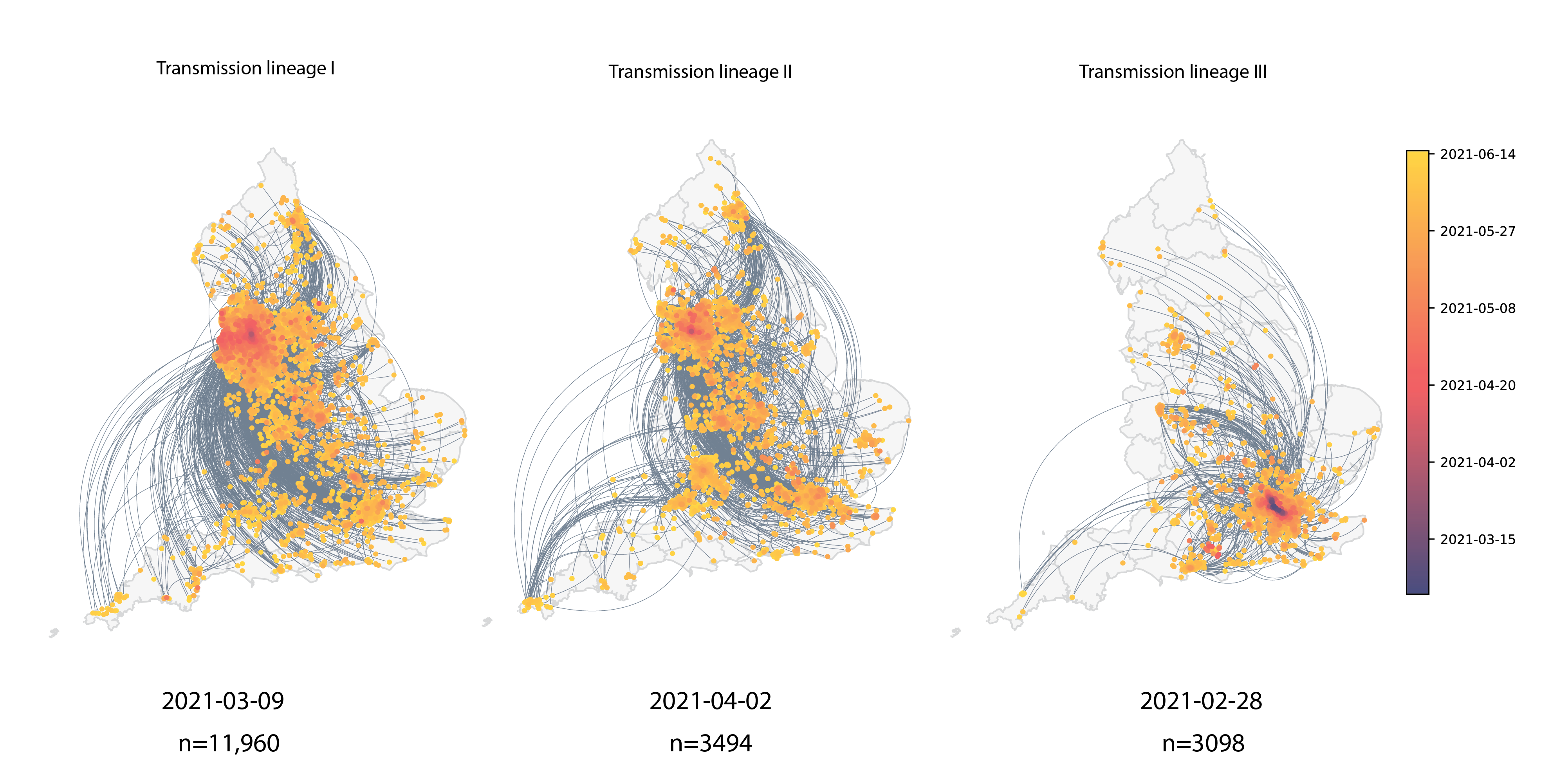
McCrone et al., 2022
Virus evolution within and between humans hosts
RNA virus evolution - Go extinct another day
- Error prone RNA dependent RNA polymerase
- High mutation rate (~10-4) mutations/nucleotide replicated (~1 mutation in every genome)
- Rapid, adaptive evolution thwarts antiviral drugs, vaccine programs, and prior immunity.

Credit: Adam Lauring
Each infection is a new roll of the dice
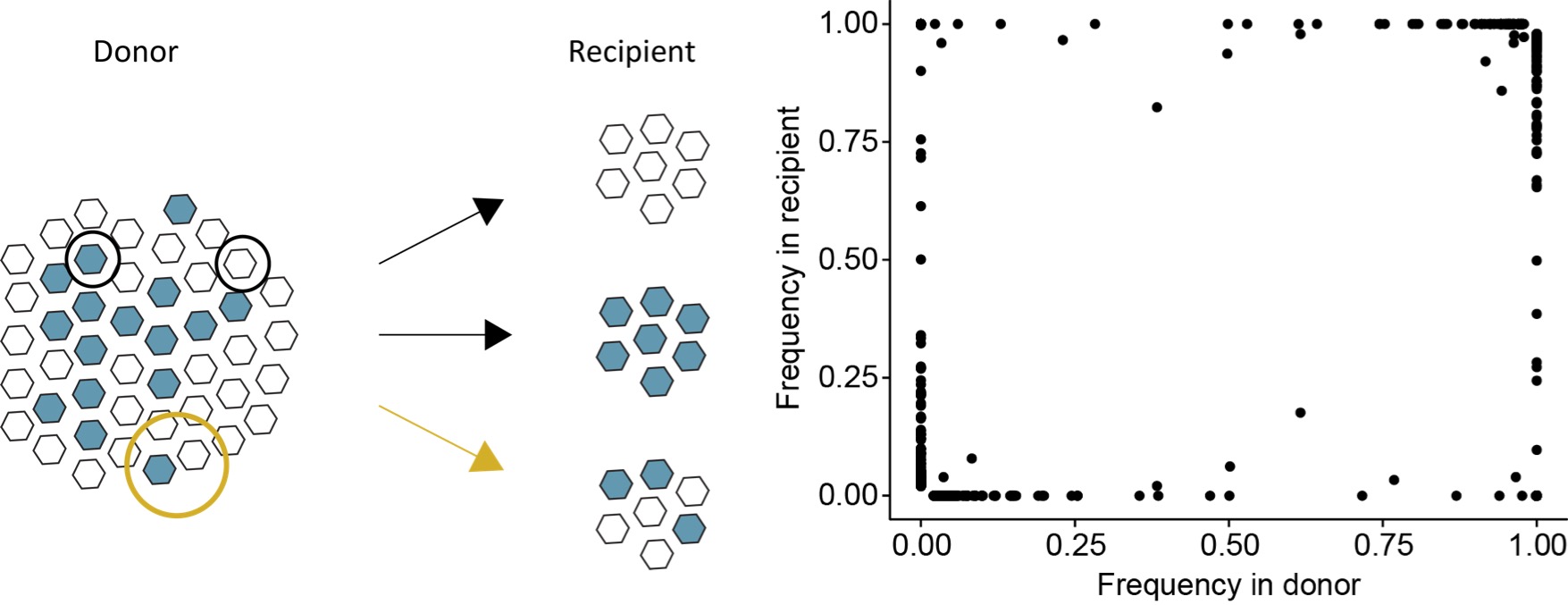
Virus evolution and transmission
Molecular clock and time trees
Assessing temporal signal
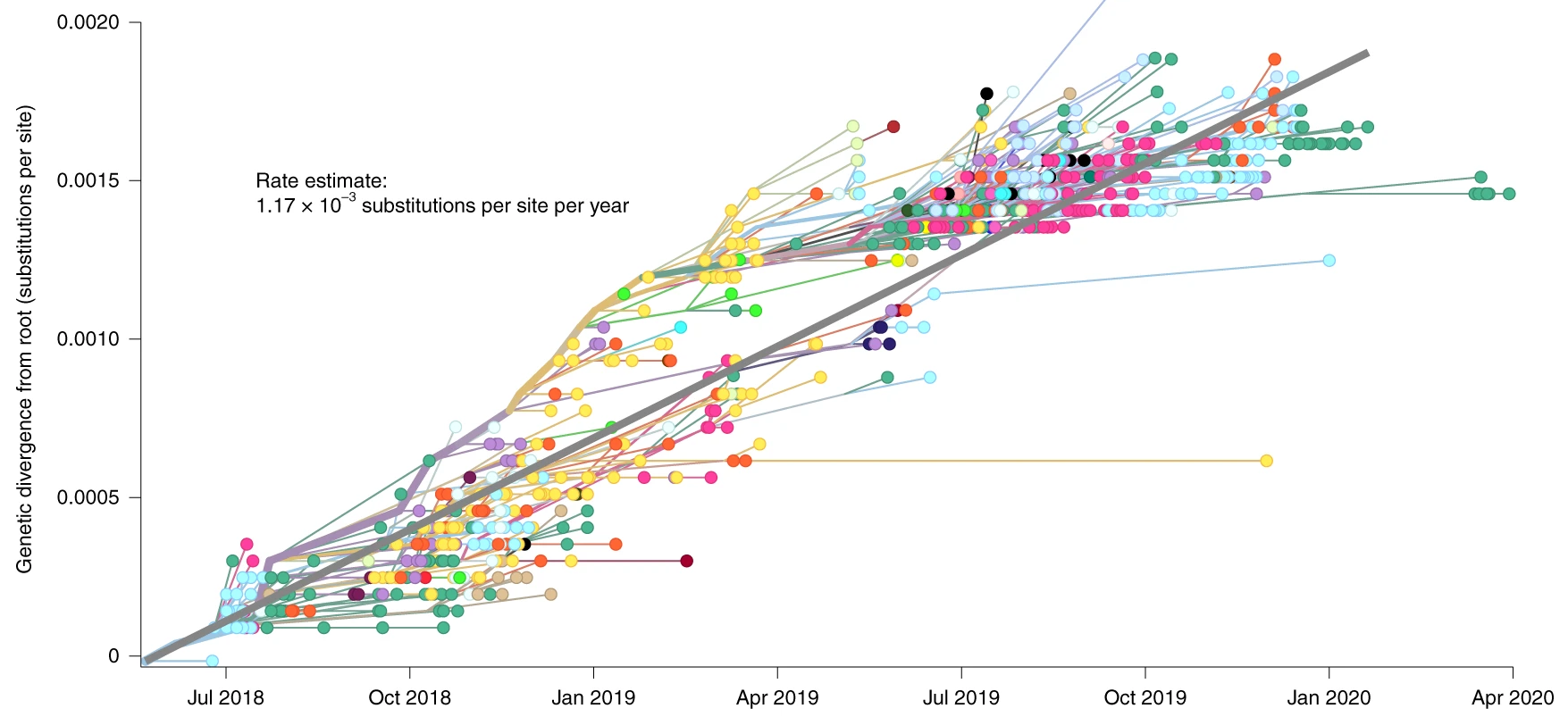
SARS-CoV-2
Sampling duration matters
Ebola virus
- (-) sense RNA virus ~18 Kb genome
- 17 independent spillover events from a poorly characterized reservoir
- Wide-spread mortality observed among great apes in Gabon and the Republic of Congo during the 1990s
- Anti-Ebola antibodies and viral RNA have been detected in multiple bat species
Ebola evolution in humans
Holmes, et al., 2016
Kinganda-Lusamaki, et al., 2021
Extreme rate heterogeneity in humans
Keita, A. K, et al., 2021
Ebola evolution 1976-2014
Geographic spread 1976-2014
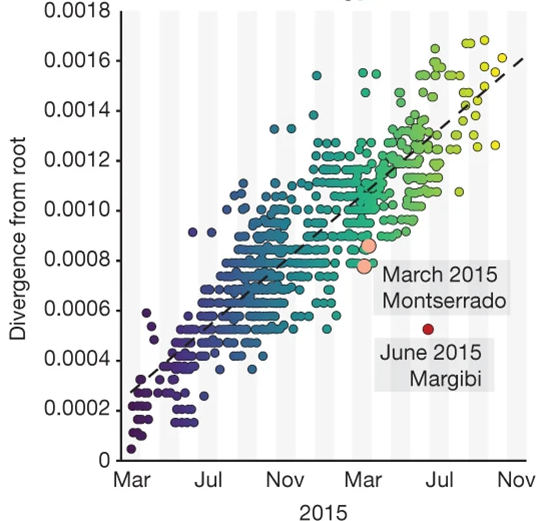
Recent outbreak are less diverged then expected
A latent branch-rate model
evolutionary rate = 0| μ
latent $ \overset{\mbox{$\lambda$}}{\Leftrightarrow}$ replicating
Evidence of latency between outbreaks
evolutionary rate = 1.52e-4
Uncertainty in the root position for up to 2014
Possible explanations
- Outside of humans ebola has not evolved according to a molecular clock
- From 1976-2014 ebola replicated/mutated at a rate significantly slower than that seen in humans
- From 1976-2014 ebola was under extreme purifying selection
- From 1976-2014 ebola replicated/mutated at a rate similar to that seen in humans
Latent branches date back to initial epidemic wave
Geographic implications of 'slow-down' events
State dependent evolution
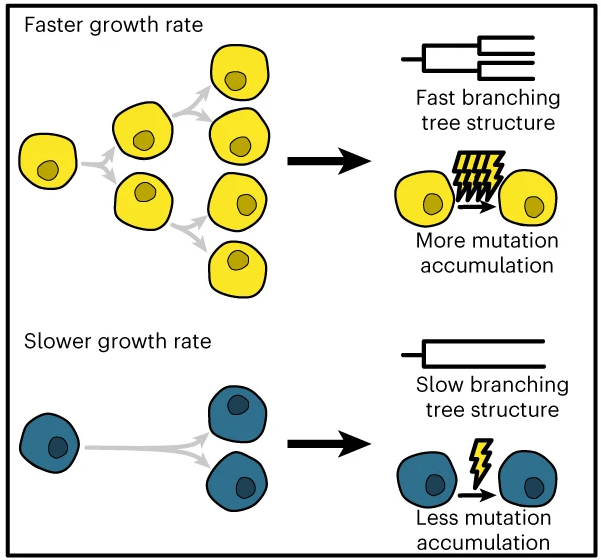
Lewinsohn, Bedford, Müller & Feder, 2023
Simulations suggest more latent lineages exist
Hypothesis: Ebola dynamics in the reservoir
- Active transmission from 1970-2014 seeding latent infections
- Recent outbreaks (2014-present) stem from reactivated infections
- Long-lived reservoir host
- Dynamic spill-over threat
- Unknown mechanism for latency

Applications to human outbreaks
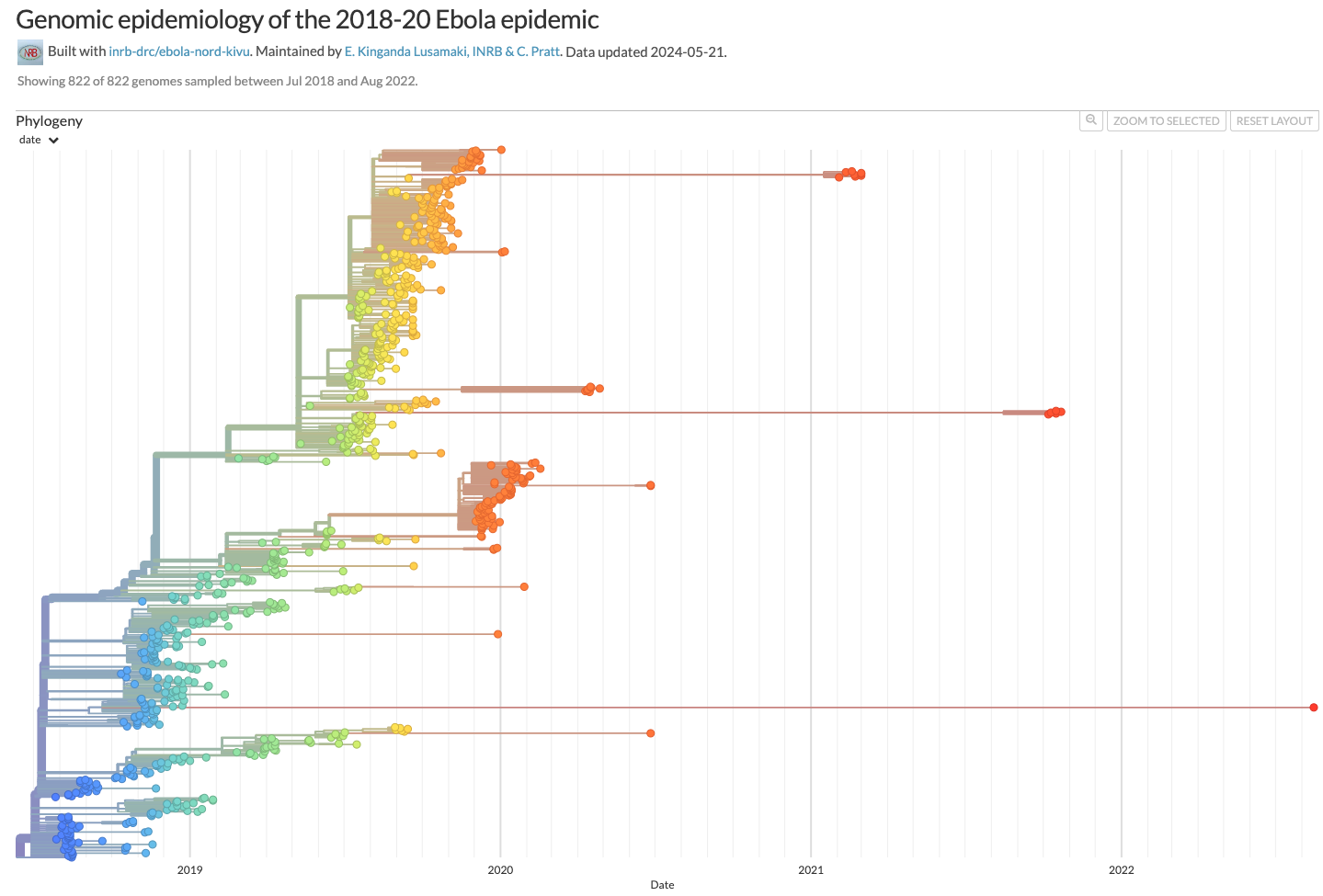
Relative risk of zoonotic vs human derived outbreaks
Thank you
University of Edinburgh
- Andrew Rambaut
- Ifeayni Omah
- Verity Hill
- Ben Jackson
- Áine O'Toole
- Rachel Colquhoun
- Emily Scher
- Shawn Yu
- Guy Baele -KU Leuven
McCrone lab
- Conner Copeland
- Cristian Ovadiuc
Bedford lab
- Nicola Muller
Matsen Group
- Eric Matsen
- Joseph Brew



