Research
RNA viruses evolve rapidly and accumulate mutations as outbreaks grow and spread. Because viral evolution is simultaneous with epidemic growth, the evolutionary relationships among sequenced infections record important information about the processes that drive epidemics. In the McCrone lab we use computational biology and phylogenetics to estimate these relationships and understand their implications for pathogen transmission.
High-resolution phylodynamics
Historically, genetic data sets have lacked the density and resolution needed for modeling pathogen spread in high detail. However, the current SARS-CoV-2 pandemic has shown that we have entered a new era of genomic epidemiology. Current and future data sets will be large enough to inform high-resolution models of viral epidemics.
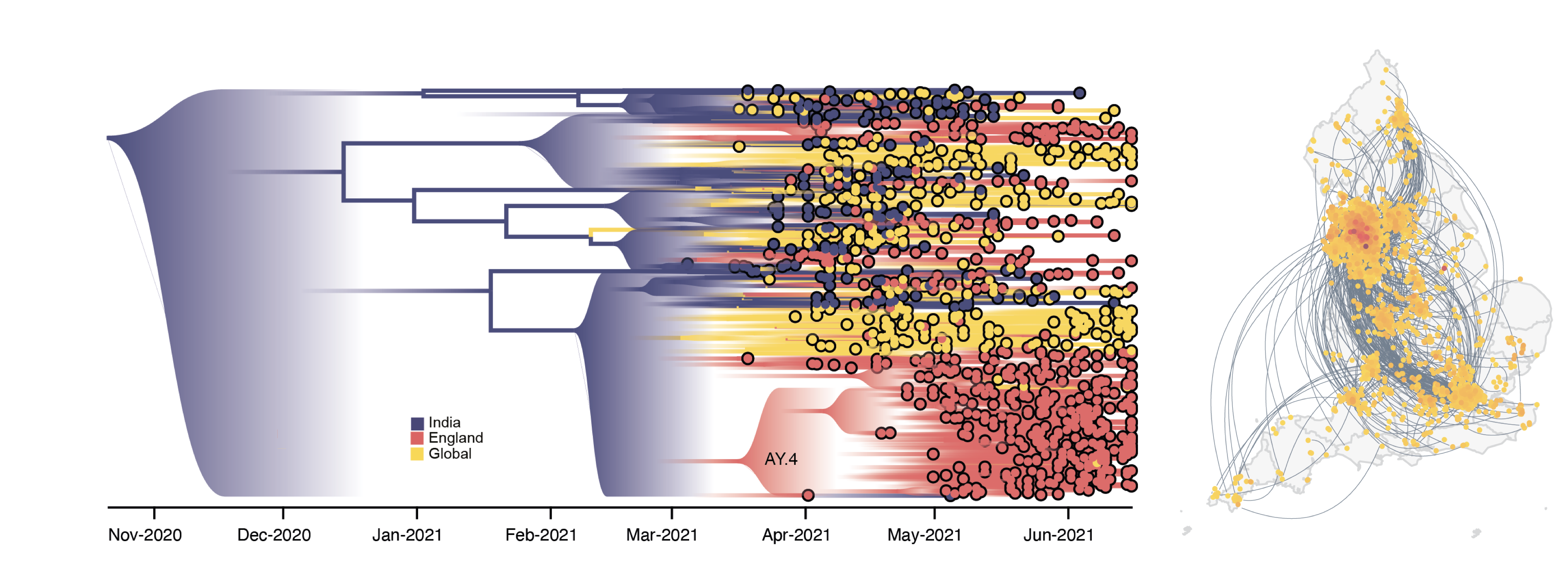
During the pandemic we were able to leverage global and local sequencing efforts to reconstruct local dynamics from genome sequences. We are working to expand this work beyond pandemic response to learn about the fundamental processes that drive transmission in different viral systems.
Novel methods for large-scale phylodynamics
Our ability to analyze tens of thousands of sequenced isolates greatly enhanced our understanding of SARS-CoV-2 evolution and spread. Such analyses are feasible because of simple models and memory efficient data structures implemented in BEAST.
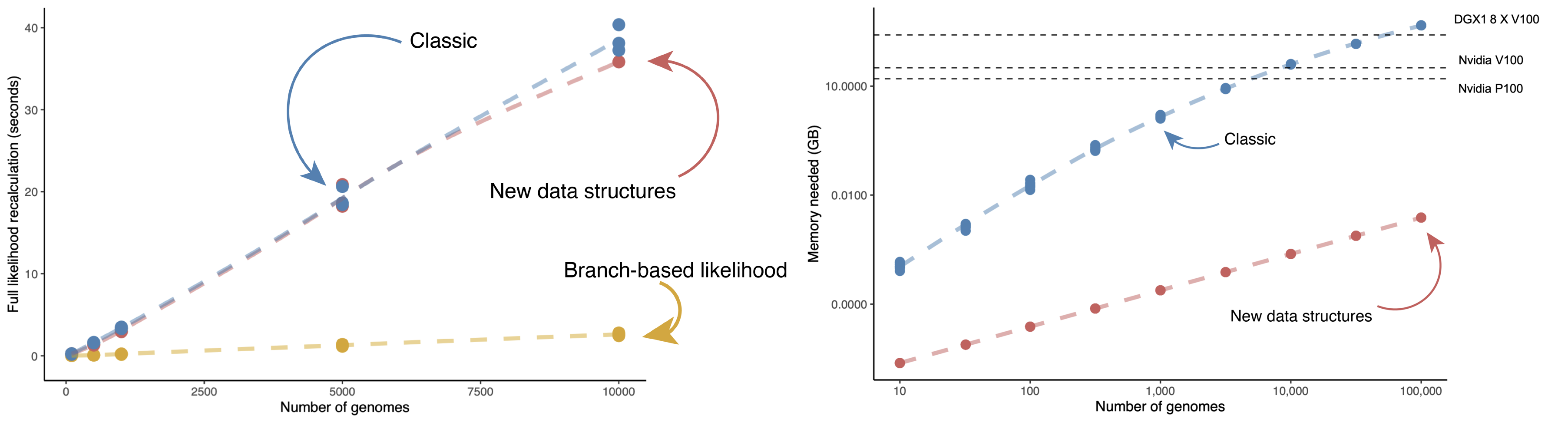
We are incorporating these models into more sophisticated statistical frameworks to answer deeper biological questions about virus evoltion and make large-scale analyses rapid, routine, and accessible.
Causes and consequences of within-host diversity
The global evolutionary dynamics of RNA viruses ultimately derive from processes that act within and between individual hosts. But within-host datasets are noisy, and we have an incomplete understanding of how tranmission bottlenecks and within-host selective pressures shape viral diveristy.
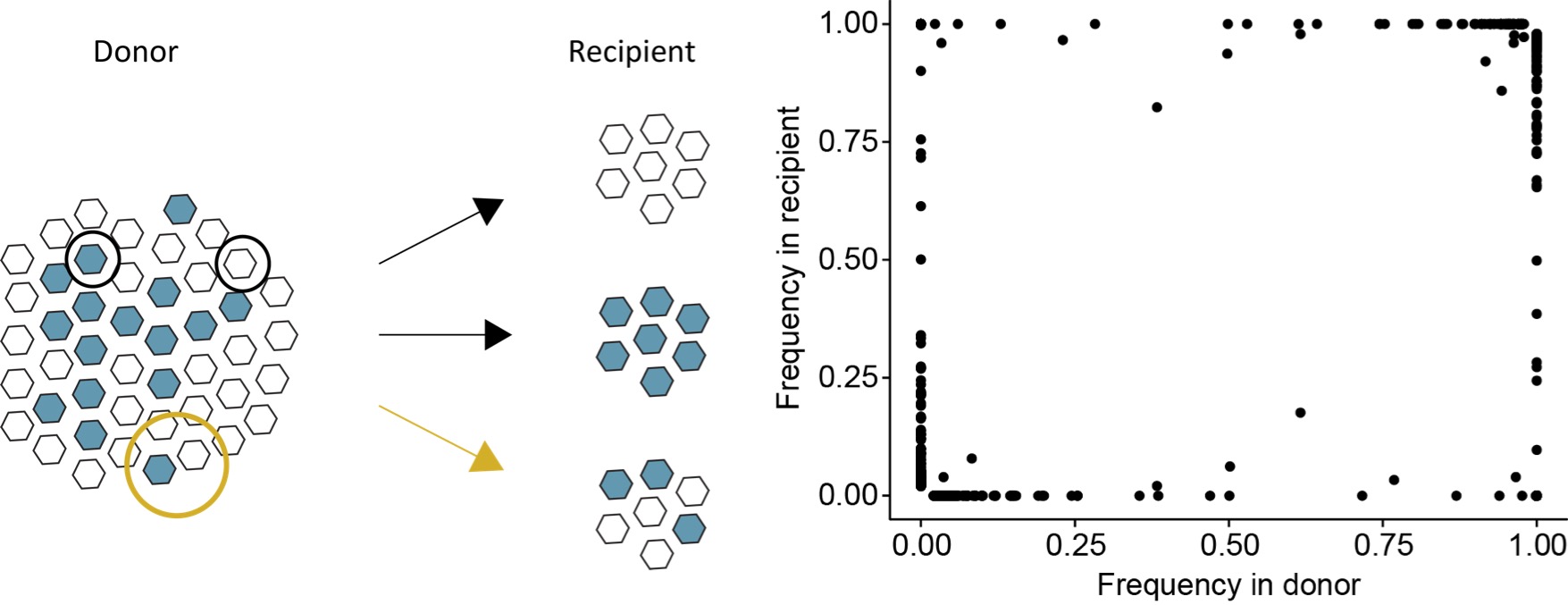
In our group we are building new models to overcome these challenges and characterize the molecular evolution of viruses at the host level.
The recent and rapid growth in sequencing capacity has the potential to revolutionize our understanding of virus evolution and epidemiology. We are working to realize that potential.